Using GTF files to extract information about genes, transcripts and related features
The Ensembl gtf file contains the comprehensive gene and transcript information for model organisms e.g. human and mouse. It can be used in RNA-Seq alignment and quantification programs such as STAR.
Downloading the appropriate GTF file
- The file can be accessed under the ‘Gene sets’ column of the table of the handy Ensembl FTP downloads website.
- for example, you can download the current versions of the mouse and human gtf files with the following commands:
curl ftp://ftp.ensembl.org/pub/release-94/gtf/mus_musculus/Mus_musculus.GRCm38.94.gtf.gz -o Mus_musculus.GRCm38.94.gtf.gz
curl ftp://ftp.ensembl.org/pub/release-94/gtf/homo_sapiens/Homo_sapiens.GRCh38.94.gtf.gz -o Homo_sapiens.GRCh38.94.gtf.gz
Formatting of the GTF file
- the GTF file is a specific type following the GFF annotation format.
- there’s actually a mixture of tabs, colons etc. used to separate fields, which you can see by viewing the file as below:
[me@XXXXX ]$ head Homo_sapiens.GRCh38.94.gtf
#!genome-build GRCh38.p12
#!genome-version GRCh38
#!genome-date 2013-12
#!genome-build-accession NCBI:GCA_000001405.27
#!genebuild-last-updated 2018-07
1 havana gene 11869 14409 . + . gene_id "ENSG00000223972"; gene_version "5"; gene_name "DDX11L1"; gene_source "havana"; gene_biotype "transcribed_unprocessed_pseudogene";
1 havana transcript 11869 14409 . + . gene_id "ENSG00000223972"; gene_version "5"; transcript_id "ENST00000456328"; transcript_version "2"; gene_name "DDX11L1"; gene_source "havana"; gene_biotype "transcribed_unprocessed_pseudogene"; transcript_name "DDX11L1-202"; transcript_source "havana"; transcript_biotype "processed_transcript"; tag "basic"; transcript_support_level "1";
Features inside the GTF file
- The third column lists the feature types, which you can also confirm with this small script from Benn:
awk '{print $3}' Homo_sapiens.GRCh38.87.gtf | sort | uniq
CDS
Selenocysteine
exon
five_prime_utr
gene
start_codon
stop_codon
three_prime_utr
transcript
- These seem to be best defined in the gtf submission guidelines under the heading ‘Attributes/Annotation features’. You’ll notice it refers to them as ‘SO’ features which actually corresponds to the Sequence Ontology.
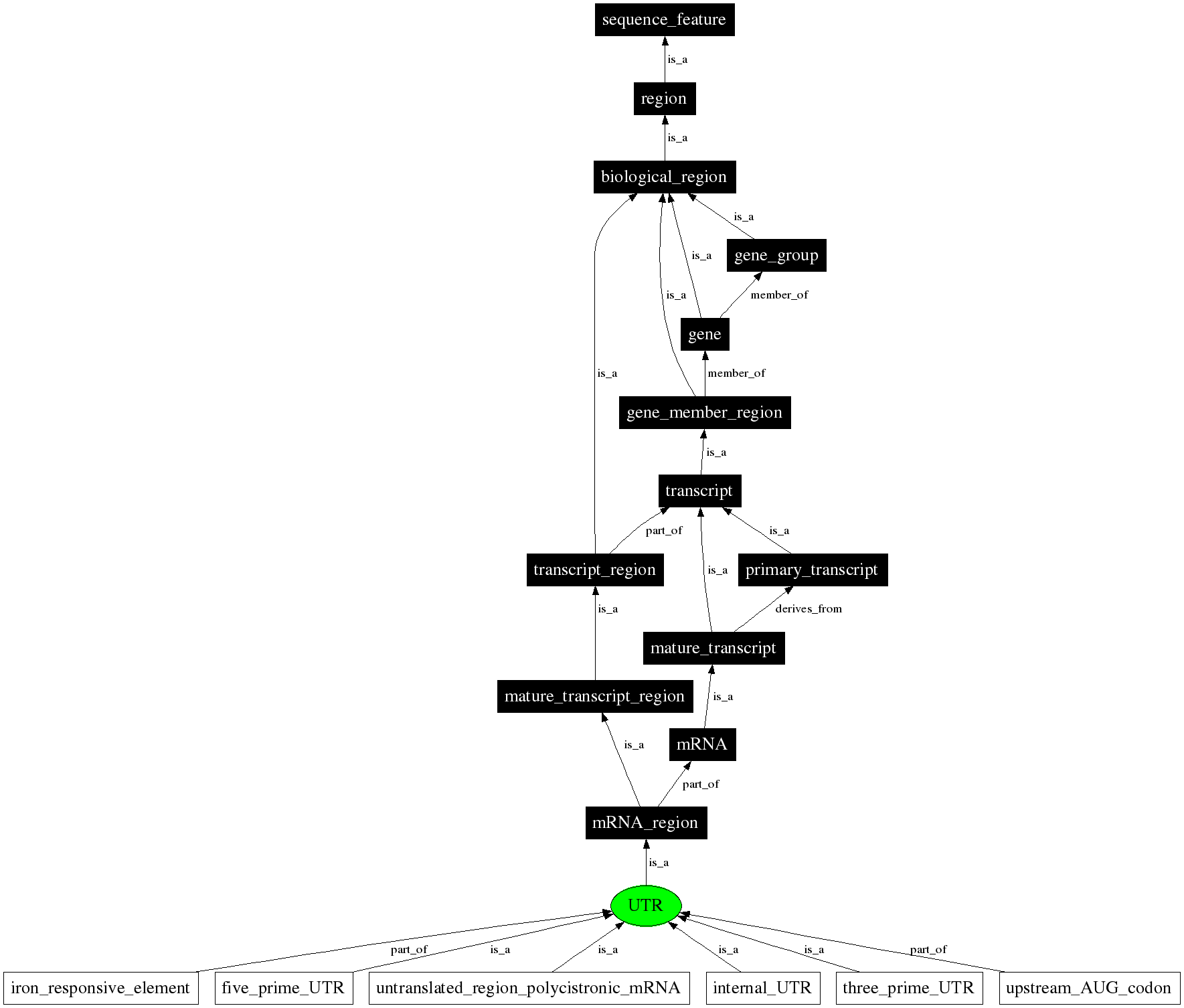
- This is also pointed out on the answer post by Istvan Albert. Indeed searching for terms gives a table containing a brief definition and the children and parents of that term which is a bit more informative. The below image shows for example that UTR is considered a portion of the mature transcript.
Extract features of interest from GTF using the command line
- The Gencode documentation has some beginner short scripts for doing this with awk within the section ‘Examples for fetching specific parts from the file’ here.
- It’s also really nicely outlined in the series of posts by Nacho Caballero here.
Import the GTF file into R
- For downstream applications, the package rtracklayer has some handy import function specifically for gtf files.
- for the given FILE_PATH you can do something like this:
# import gtf file -------------------------------------
mm_gtf <- rtracklayer::import('FILE_PATH/Mus_musculus.GRCm38.94.gtf')
head(mm_gtf)
# extract gene features from the gtf -------------------------------------
unique(levels(mm_gtf$type))
genes_only <- mm_gtf[mm_gtf$type == "gene",]
head(genes_only)
Written on June 16, 2019
